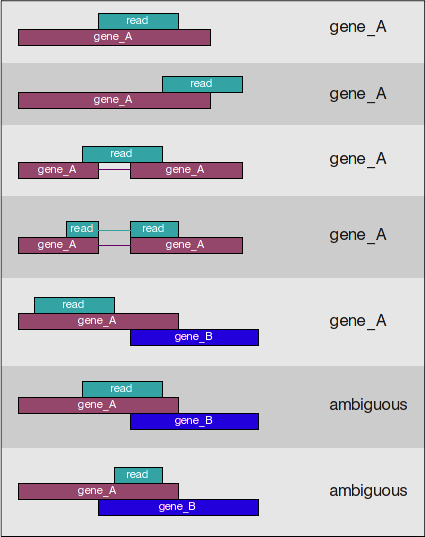
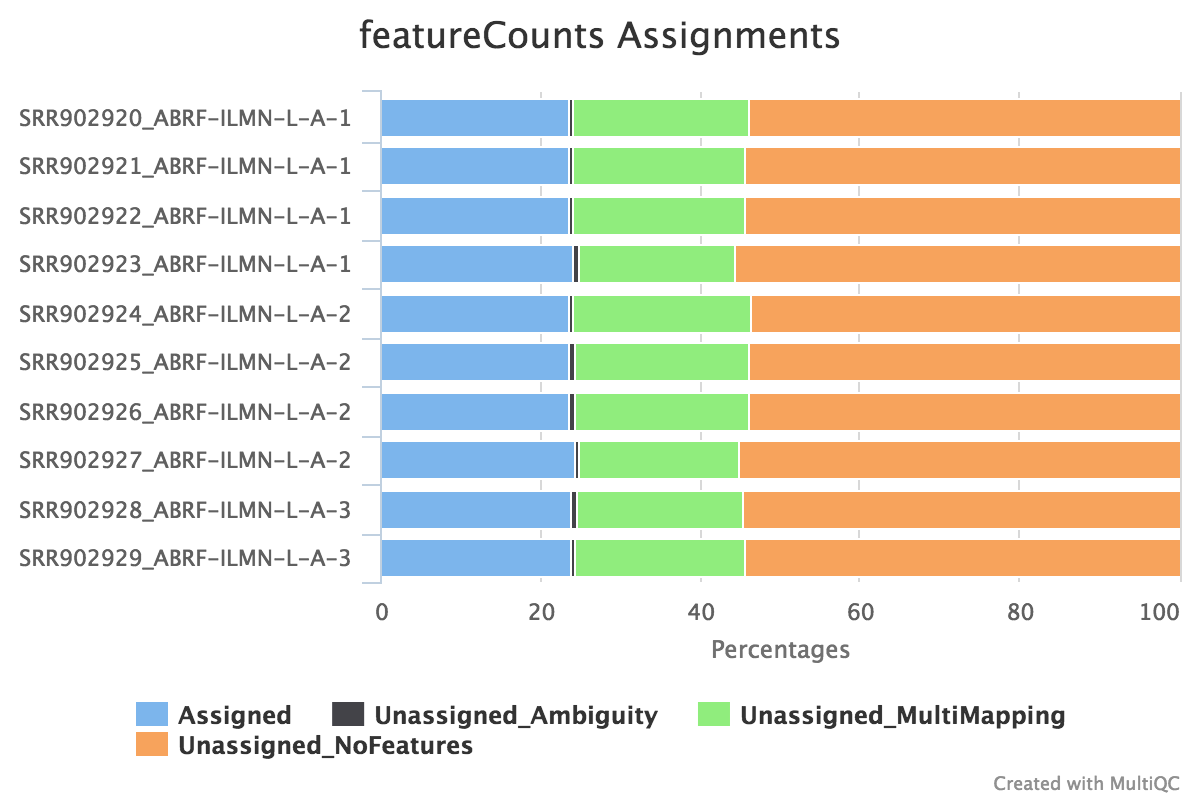
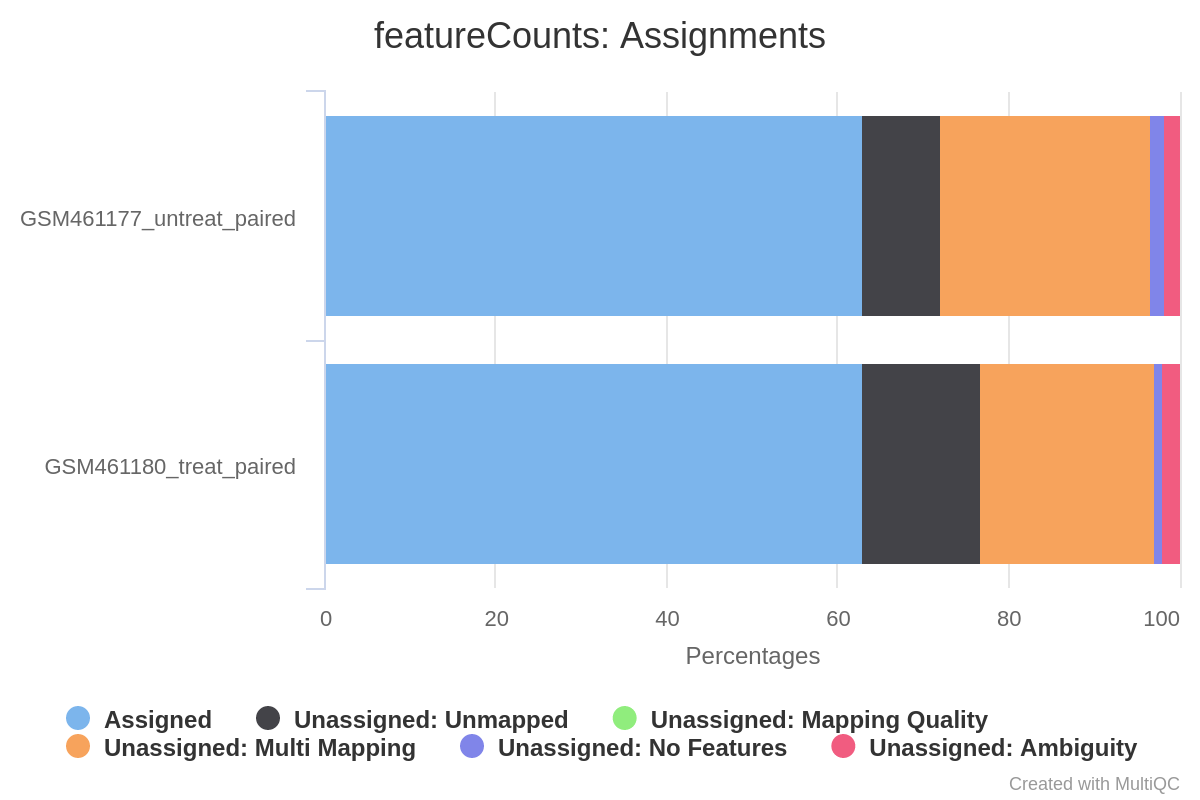
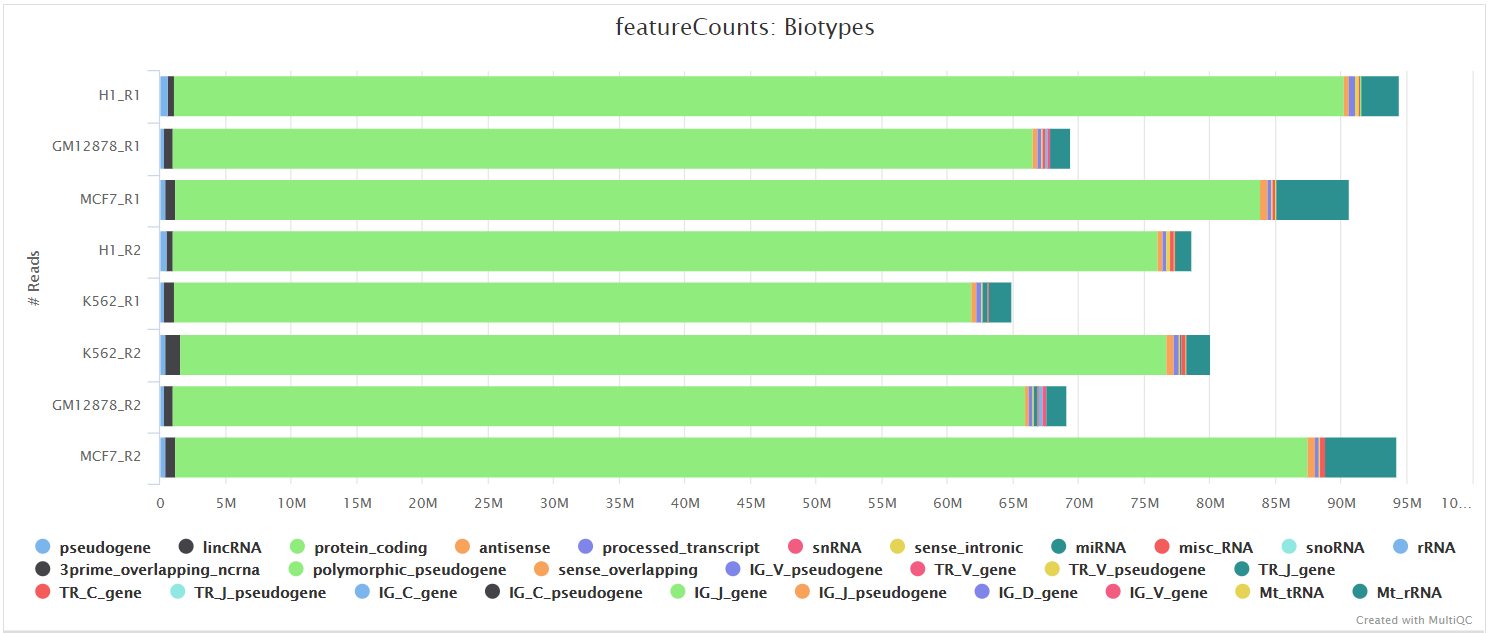
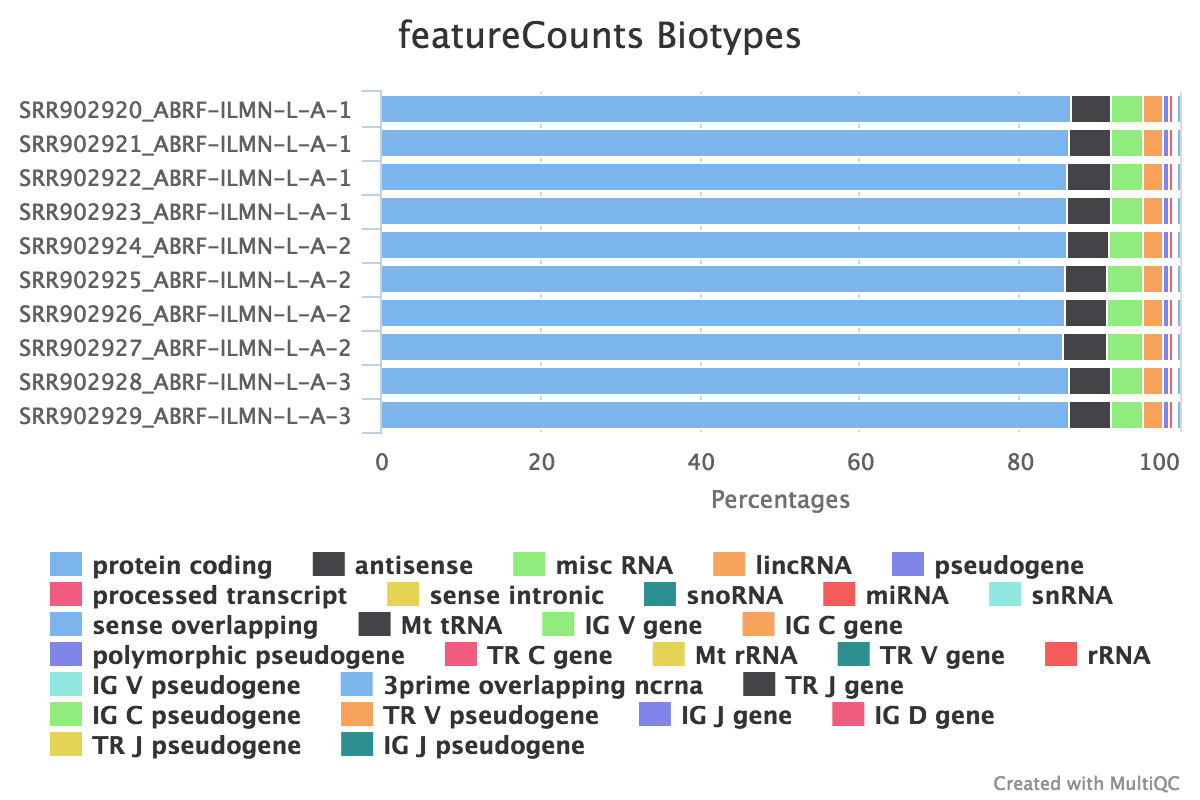
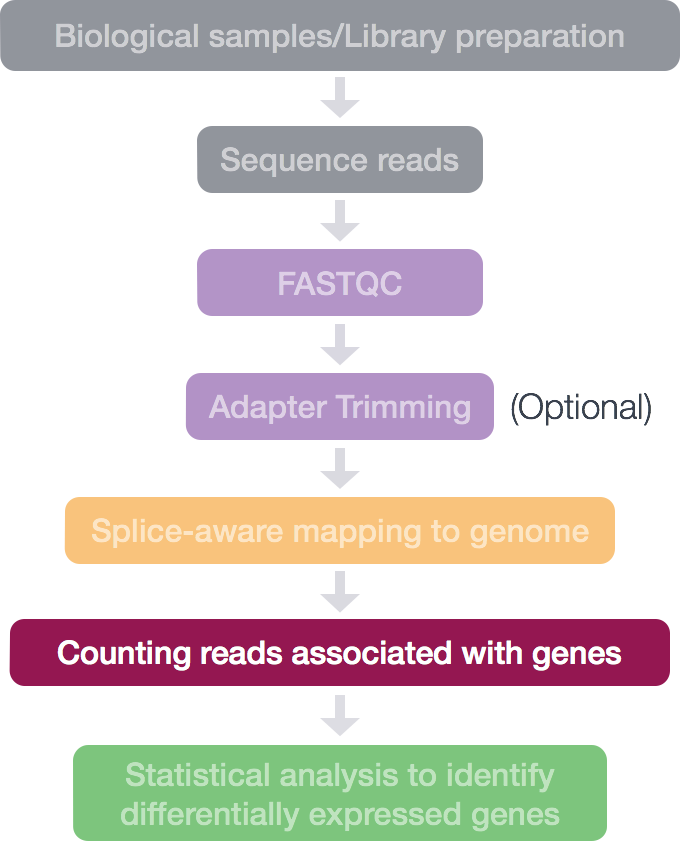
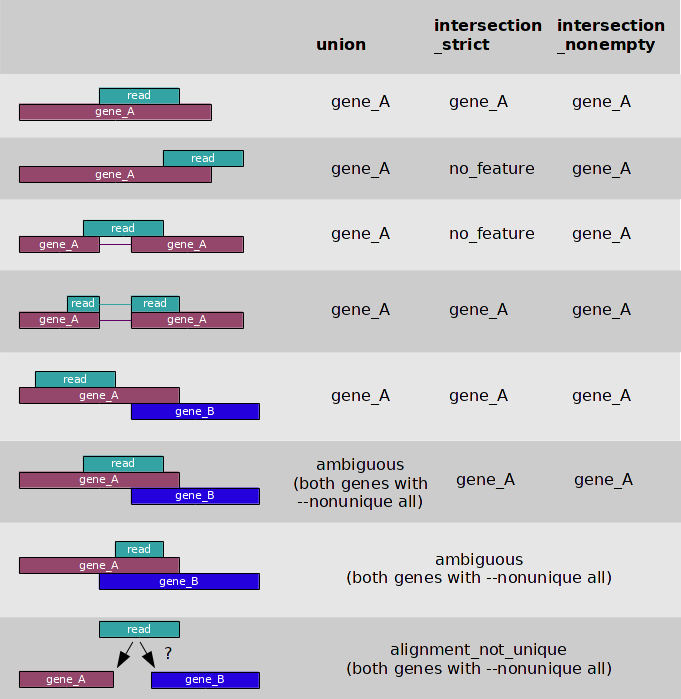
Federico Giorgi on X: "#RNASeq: what happens to multi-mapping reads? Whoah apparently #htseq and #featureCounts ignore them by default! #Salmon, #Sailfish and #Kallisto are the way, with the Expectation Maximization algorithm! @LabMutwil

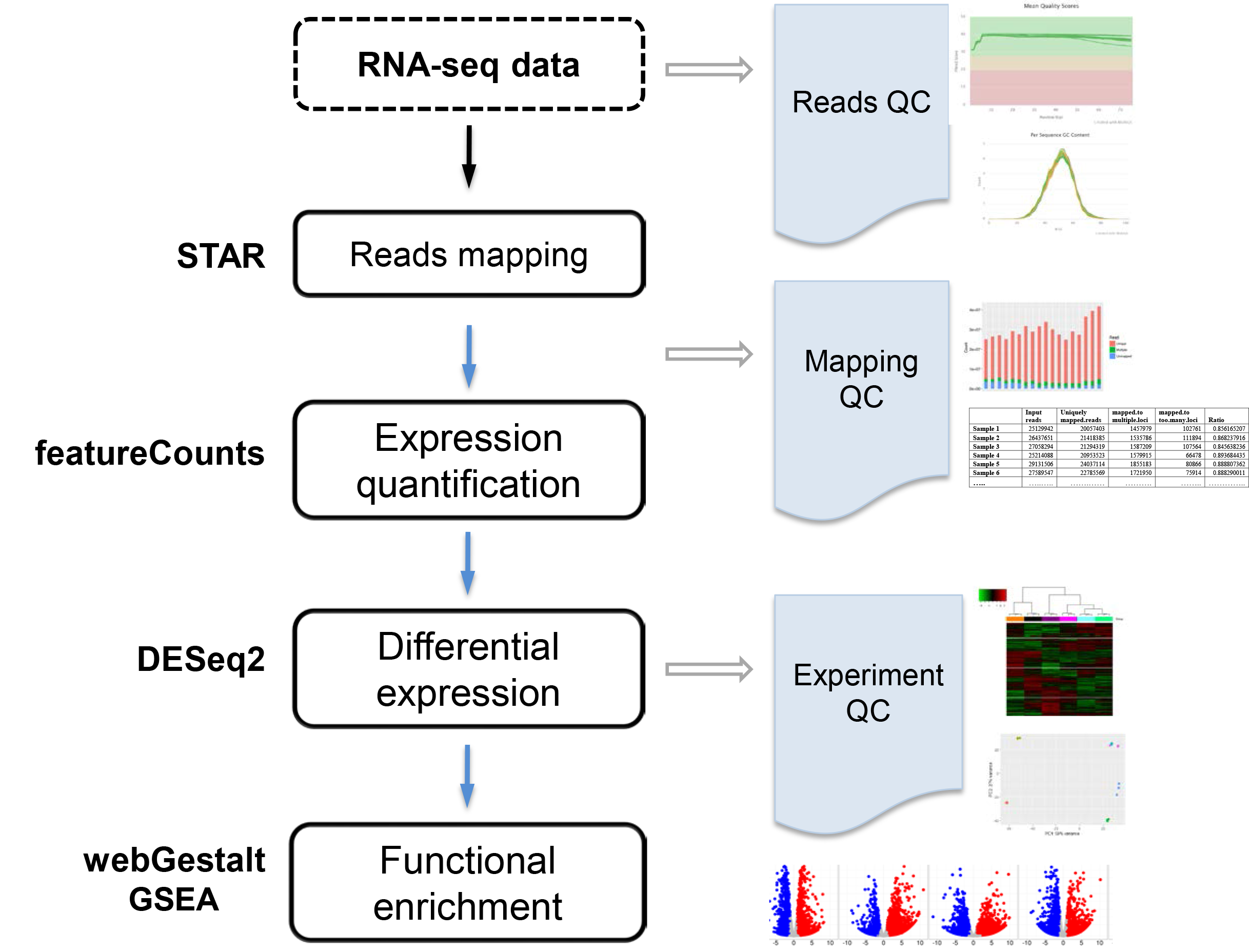
Statistics generated using Gene Counts mode of alignment tool STAR. The... | Download Scientific Diagram

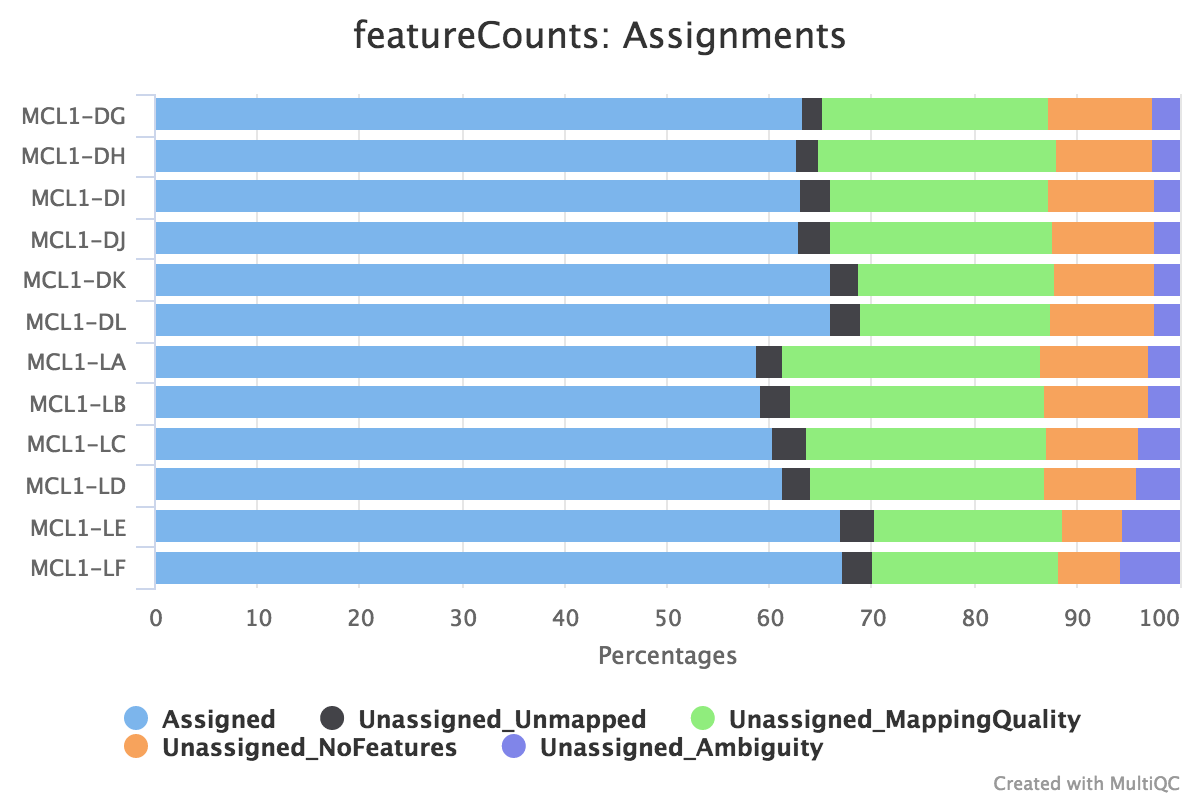
below 50 percents of reads are assigned in featurecount - usegalaxy.org support - Galaxy Community Help

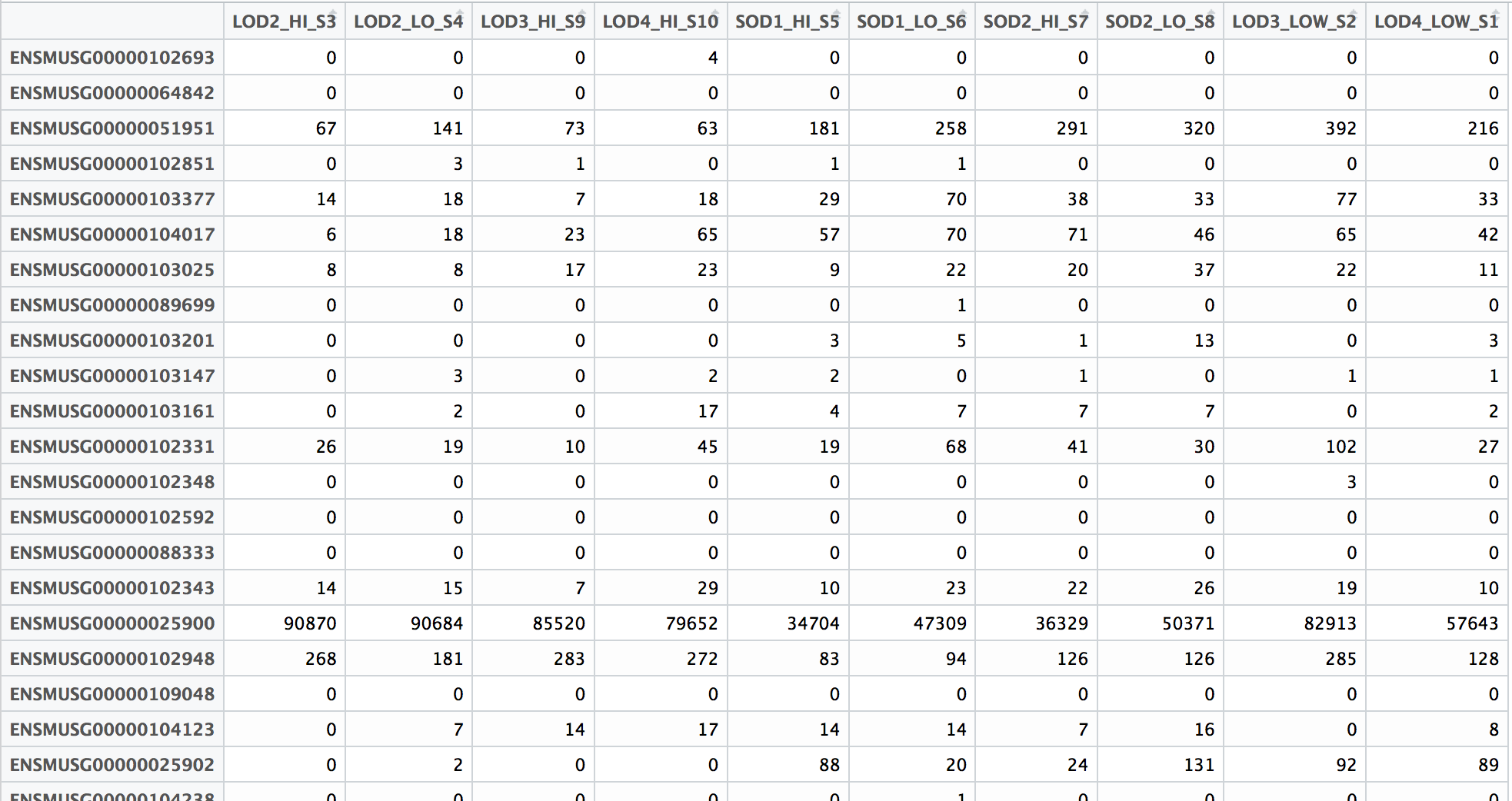
Compute the number of reads mapped to genomic features - MATLAB featurecount - MathWorks Deutschland